The GENESIS simulator is undergoing a major redevelopment effort that will result in GENESIS version 3.
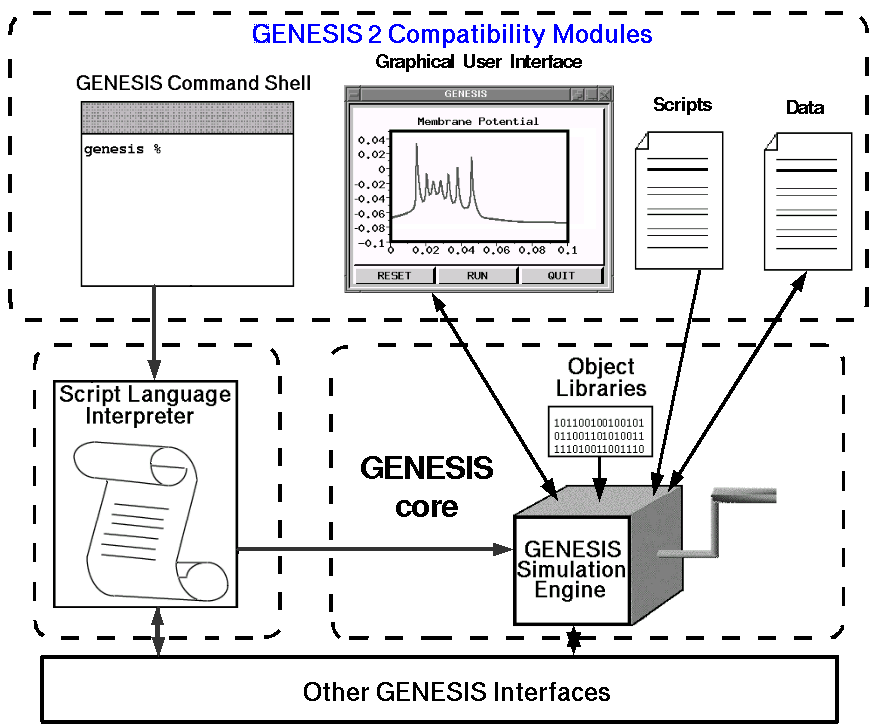
The diagram below shows the components of GENESIS 2, as seen by the GENESIS user. Although GENESIS and its scripting language produce modular object-oriented simulations that are easy to modify and extend, this is not the case with the source code for GENESIS 2. The core modules of GENESIS were written in the late 1980's in C, and the modules are much more tightly coupled than the diagram would suggest. Although the simulation code is very efficient, it is difficult to add more modern Java-based graphical interfaces, alternate script parsers, and interfaces via the WWW.

In order to provide greater interoperability with other neuroscience software tools, databases, and modeling environments, we are modularizing the GENESIS architecture, as indicated by the dashed lines in the figure above. The modular architecture that is being developed has the advantages of
This provides three significant advantages for the software system:
1) Modules can be run separately on different machines. For example, the GUI and modeling environment might run locally, while the simulator is run elsewhere either serially or in parallel on more powerful machines.
2) Decomposition of an application into multiple software components allows reuse and extension of individual modules, whether stand alone or otherwise, clearly facilitating model development and research progress.
3) Individual components can be independently updated, enhanced, or replaced when needed, thus the life cycle of a modular architecture is smoother than that of a non-scalable application.
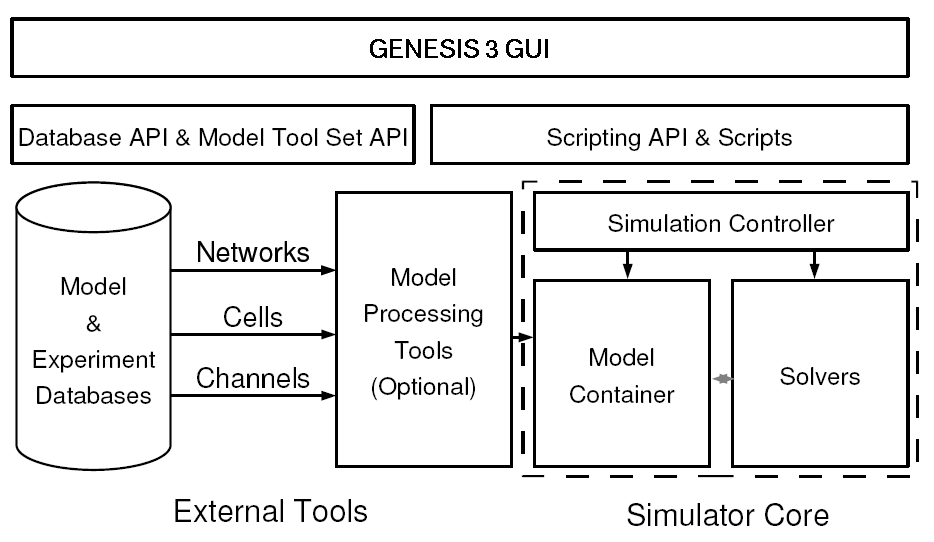
The core components of the architecture are shown below. On the bottom left are databases of neuronal models or experimental data that can be accessed by the simulator. Optional model processors (e.g. the reconstruct interface) load a model into the model container. The model container translates biological concepts and properties into mathematical formulae that can be understood by numerical solvers. Thus the numerical solvers are indepent of the biological model representation. The simulation controller orchestrates and synchronizes the actions taken by the model container (e.g. when to load a model, what stimulation to apply, when to export a model) and solvers (when to fetch the model from the model container, when to start the calculations, and what the output variables are).
The scripting layer allows the simulation system to be driven from multiple scripting languages. Python and Perl are currently supported, and for backwards compatibility, the GENESIS 2 SLI is being incorporated. The GENESIS 3 GUI is shown at the top. It allows one to import models from databases or construct them from scratch, explore model structure and parameters, and visualize variables and behavior.
The core modules for GENESIS 3.0 that have already been implemented and tested include: (1) Neurospaces, a declarative model container that generates highly efficient data structures implementing biological structures and components in a uniform data model across different levels of scale, which when linked to a user interface provides an efficient way to construct, inspect, and validate models, (2) Heccer, a fast compartmental solver based on the refactoring of the GENESIS compartmental solver, hsolve. The design flexibility of the architecture allows Heccer to be instantiated from C, Perl, or other scripting languages, (3) SSP (Simple Scheduler in Perl), a simulation control file used to bind Neurospaces and Heccer for testing and development purposes, which is now serving as a template to build a more complex scheduler, and (4) MOOSE, which reimplements parts of the GENESIS mathematical solvers for kinetic modeling and simulation, GNU Scientific Library ODE solvers, and transparent parallelization. Validation of MOOSE for GENESIS will allow it to replace SSP.
There are two ongoing projects that are developing a GENESIS-compatible replacement for the GENESIS core.
Hugo Cornelis (Bower lab, UT San Antonio), who programmed many of the advanced features of the GENESIS hsolve object, is developing Neurospaces and its compartmental model solver Heccer. This promises to be extremely efficient for the solution of large compartmental neuron models. For more information about Neurospaces and Heccer, see http://www.neurospaces.org.
Upinder Bhalla (NCBS Bangalore), who wrote the X Windows XODUS GUI interface for GENESIS and did much of the initial GENESIS development, has been rewriting the GENESIS base code in C++, as MOOSE. This reimplementation in an object-oriented language makes it simple to "unbundle" the components of GENESIS.
The following outline summarizes MOOSE. More information may be found at http://moose.sourceforge.net/.
The Messaging Object Oriented Simulation Environment (MOOSE) provides:
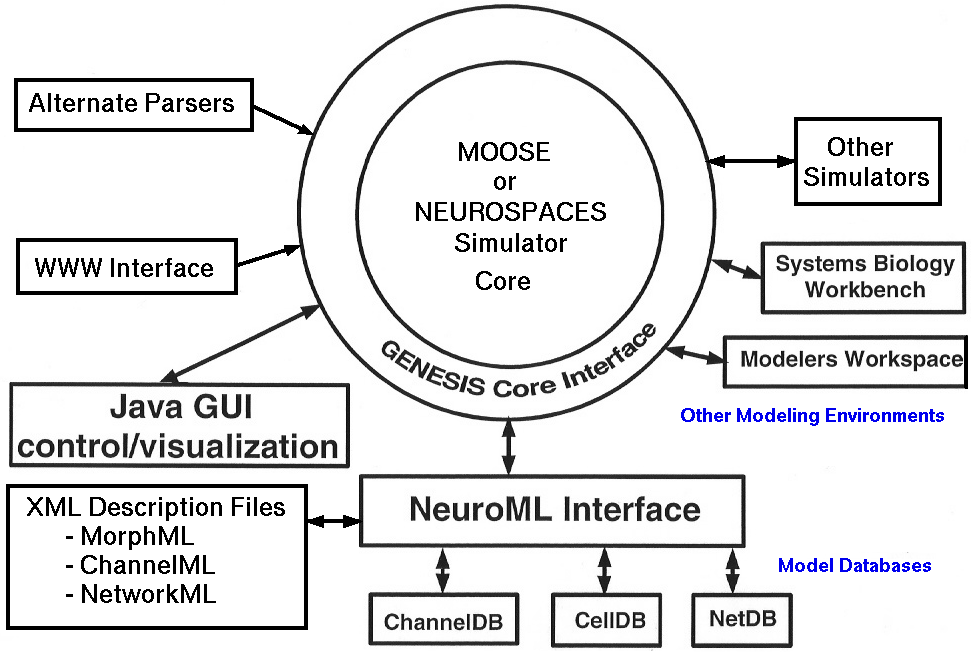
Using this architecture, G3 is being developed with the necessary interfaces that will, in principle, allow any simulation system to use its features. The last figure shows the multiple external interfaces to GENESIS 3 that will be allowed with the new architecture.
Some of the current priorities for GENESIS 3 development are to: