To appear in The Handbook of Brain Theory and Neural Networks, Second
edition, (M.A. Arbib, Ed.), Cambridge, MA: The MIT Press, 2002.
http://mitpress.mit.edu ©The MIT Press
James M. Bower
bower@uthscsa.edu
Research Imaging Center, University of Texas Health Science Center
and Cajal Neuroscience Research Center, University of Texas
San Antonio, TX 78284
David Beeman
dbeeman@dogstar.colorado.edu
Dept. of Electrical and Computer Engineering
University of Colorado
Boulder, CO 80309
Michael Hucka
mhucka@caltech.edu
Control and Dynamical Systems 107-81
California Institute of Technology
Pasadena, CA 91125
GENESIS (the GEneral NEural SImulation System) was developed as a research tool to provide a standard and flexible means for constructing structurally realistic models of biological neural systems. ``Structurally realistic'' simulations are computer-based implementations of models whose primary objective is to capture what is known about the anatomical structure and physiological characteristics of the neural system of interest. The GENESIS project is based on the belief that progress in understanding structure-function relationships in the nervous system specifically, or in biology in general, will increasingly require the development and use of structurally realistic models (Bower, 1995). It is our view that only through this type of modeling will general principles of neural or biological function emerge.
There is considerable debate within the computational neuroscience community concerning the appropriate level of modeling. As illustrated in other chapters in this volume, many modeling efforts are currently focused on abstract ``general'' representations of neural function rather than detailed, realistic models. However, the history of science clearly indicates that realistic models play an essential role in the development of quantitative understandings of physical systems. For example, philosophers and priests for thousands of years invented ``models'' to account for the motion of the planets in the night sky. These models, the most famous of which is probably the Ptolemeic system, ``replicated the data'' and made quantitative predictions. The structure of these planetary models, however, already assumed the general principles on which the universe was organized. It was not until the 16th and 17th centuries when Kepler and later Newton constructed realistic models of the solar system that general principles such as universal gravitation emerged. The inverse square law for gravitational attraction fell out of a model that Newton constructed of the moon's movement around the earth; it was not an apple-induced inspiration.
It is our view that neuroscience is not yet ready for its Newton. Instead, we are still in need of Kepler. Viewed most generally, GENESIS is intended to provide a framework for quantifying the physical description of the nervous system in a way that promotes common understanding of its physical structure. At the same time, this physical description also provides the base for simulations intent on understanding fundamental relationships between the structure of the brain and its measurable behavior. Again, looking back at the evolution of planetary science, Kepler's realization that the motion of the planets was eliptical came about as a result of his careful analysis of the detailed positions of the planets obtained by Tycho Brahe. Kepler's development of a mathematical formalism to describe eliptical motion provided a seminal framework for the work of later physicists including Newton. Similarly, it is our hope and expectation that the formalism being developed within the GENESIS project and other simulation systems such as NEURON (see THE NEURON SIMULATION ENVIRONMENT) will provide a means for neurobiologists to collaboratively construct a physical description of the nervous system. We believe strongly that general principles of organization, function, and computation will only emerge once this description has been constructed.
GENESIS was designed from the beginning to allow the development of simulations at any level of complexity, from sub-cellular components and biochemical reactions, to whole cells, networks of cells and systems-level models. The earliest GENESIS simulations were biologically realistic large-scale simulations of cortical networks (Wilson and Bower, 1992). The De Schutter and Bower (1994a,b) cerebellar Purkinje cell model is typical of a large detailed single-cell model, with 4550 compartments and 8021 ionic conductances. GENESIS is now being used for large systems-level models of cerebellar pathways (Stricanne, Morissette and Bower, 1998), and at the other extreme, is increasingly being used to relate cellular and network properties to biochemical signaling pathways (Bhalla and Iyengar, 1999).
Although GENESIS continues to be widely used for single cell modeling, and for modeling small networks (for example, see HALF-CENTER OSCILLATIORS UNDERLYING RHYTHMIC MOVEMENTS), we have seen in recent years a dramatic increase in the number of publications that report the use of GENESIS for large network models. We believe that this trend is largely due to the availability of our libraries of ion channels and complete cell models. A description of some notable large-scale network GENESIS simulations (many using parallel computers) that have been published recently may be found at http://www.genesis-sim.org/GENESIS/research/genres.html. This web page also contains links to a list of 170 papers based on research with GENESIS from groups outside of Caltech, and a summary of what various research groups are doing with GENESIS.
GENESIS is implemented in C, using the X Window System, and runs under most varieties of UNIX, including Linux. There is also a parallel version of GENESIS (called PGENESIS) that runs on workstation networks, small-scale parallel computers and large, massively-parallel supercomputers. PGENESIS is being used for simulations that must be run many times independently (e.g., parameter searches), and for large scale models (especially network models with thousands of neurons).
The objectives of this project were to reduce redundant software design efforts, establish standards for simulation technology, and provide a common base for the exchange of models and scientific information. The object-oriented nature of the software allows different modelers to easily exchange and reuse whole models or model components. GENESIS also includes a customizable user interface for use by modelers and educators. From the beginning, GENESIS was also designed to serve as an instructional tool, because our involvement in several educational projects has demonstrated that simulations can provide flexible and dynamic learning tools for neuroscience education (Bower and Beeman, 1998).
The design of the GENESIS simulator and interface is based on a ``building block'' approach. Simulations are constructed from modules that receive inputs, perform calculations on them, and then generate outputs. Model neurons are constructed from these basic components, such as compartments (short sections of cellular membrane) and variable conductance ion channels. Compartments are linked to their channels and are then linked together to form multi-compartmental neurons of any desired level of complexity. Neurons may be linked together to form neural circuits. This object-oriented approach is central to the generality and flexibility of the system, as it allows modelers to easily exchange and reuse models or model components. In addition, it makes it possible to extend the functionality of GENESIS by adding new commands or simulation components to the simulator, without having to modify the GENESIS base code.
Neural systems are particularly amenable to this object-oriented approach because they typically consist of discrete components interacting in quite stereotyped ways, and because the different simulations tend to use similar neural components, display routines, numerical integration routines, etc. This modularity means that it is possible to quickly construct a new simulation or to modify an existing simulation by changing modules that are chosen from a library or database of standard simulation components. Individual modules or linked assemblies of modules (such as compartments with channels, entire cells, or networks of cells) may be easily replicated.
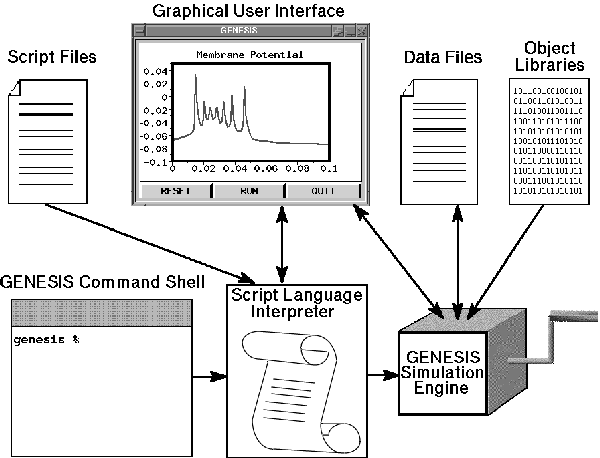
GENESIS uses a high-level simulation language to construct neurons and their networks. Commands may be issued either interactively to a command prompt, by use of simulation scripts, or through the graphical interface. A particular simulation is set up by writing a sequence of commands in the scripting language that creates the network itself and the graphical interface for a particular simulation. The scripting language and the modules are powerful enough that only a few lines of script are needed to specify a sophisticated simulation. The principal components of the simulation system and the various modes of interacting with GENESIS are illustrated in Fig. 1.
The underlying level of the GENESIS user interface is the Script Language Interpreter (SLI). This is a command interpreter similar to a Unix system shell with an extensive set of commands related to building, monitoring and controlling simulations. GENESIS simulation objects and graphical objects are linked together using the scripting language. The interpreter can read SLI commands either interactively from the keyboard (allowing interactive debugging, inspection, and control of the simulation), or from files containing simulation scripts.
The graphical user interface (GUI) is XODUS, the X-windows Output and Display Utility for Simulations. This provides a higher level and user-friendly means for developing simulations and monitoring their execution. XODUS consists of a set of graphical objects that are the same as the computational modules from the user's point of view, except that they perform graphical functions. As with the computational modules, XODUS modules can be set up in any manner that the user chooses to display or enter data. Furthermore, the graphical modules can call functions from the script language, so the full power of the SLI is available through the graphical interface. This makes it possible to interactively change simulation parameters in real time to directly observe the effects of parameter variations. For example, the mouse may be used to plant recording or injection electrodes into a graphical representation of the cell. In addition to provisions for plotting the usual quantities of interest (membrane potentials, channel conductances, etc.), XODUS has visualization features that permit such things as using color to display the propagation of action potentials or other variables throughout a multi-compartmental model, and to display connections and cell activity in a network model.
The ``GENESIS Simulation Engine'' consists of the simulator base code that provides the common control and support routines for the system, including those for input/output and for the numerical solution of the differential equations obeyed by the various neural simulation objects. GENESIS provides a choice of numerical integration methods, including highly accurate and stable implicit methods such as Crank-Nicholson (De Schutter and Beeman, 1998).
In addition to receiving commands from the SLI and the GUI, the simulation engine can construct simulations using information from data files and from the pre-compiled GENESIS object libraries. For example, the GENESIS ``cell reader'' allows one to build complex model neurons by reading their specifications from a data file, instead of from a lengthy series of GENESIS commands delivered to the SLI. Similarly, network connection specifications may be read from a data file with the ``fileconnect'' command.
The GENESIS object libraries contain the building blocks from which many different simulations can be constructed. These include the spherical and cylindrical compartments from which the physical structure of neurons are constructed, voltage and/or concentration activated channels, dendro-dendritic channels, and synaptically-activated channels with synapses of several types including Hebbian and facilitating synapses. In addition, there are objects for computing intracellular ionic concentrations from channel currents, for modeling the diffusion of ions within cells, and for allowing ligand gating of ion channels. There are also a number of ``device objects'' that may be interfaced to the simulation to provide various types of input to the simulation (pulse and spike generators, voltage clamp circuitry, etc.) or measurements (peristimulus and interspike interval histograms, spike frequency measurements, auto- and cross-correlation histograms, etc.).
The kinetics library supports kinetic-level modeling of biochemical pathways. This library currently includes objects for pools (molecular components), n-molecular reactions, enzymes, and channels to couple pools of different volume. The parameter search library provides a collection of objects and functions that automate the tedious process of adjusting model parameters to best reproduce experimental measurements carried out on the sytem being modeled.

Figure 1: The components of GENESIS and modes of interaction. The Script Language Interpreter processes commands entered through the keyboard, script files, or the Graphical User Interface, and passes them to the GENESIS Simulation Engine. The Simulation Engine also loads compiled object libraries, reads and writes data files, and interacts with the GUI.
In addition to the object libraries that are compiled into GENESIS, there are a number of libraries that are implemented as simulation scripts. These are available within the GENESIS distribution and in the archives of the GENESIS Users Group.
The channel library currently contains models for 39 different types of potassium channels, including several types of calcium-dependent channels, 24 types of sodium channels and 14 types of calcium channels. The available single cell models include cerebral cortical pyramidal cells, hippocampal pyramidal cells, cerebellar Purkinje cells, mitral, granule, and tufted cells from the olfactory bulb, a hippocampal granule cell model, a thalamic relay cell, and an Aplysia R15 bursting pacemaker cell.
GENESIS makes use of the GUI to provide other features to make the simulator more easily useable by people with limited programming experience. A set of ``kits'', implemented as simulation scripts, has been provided to ease the modeling process. Neurokit provides an environment for building, modifying and testing single cell models without any programming on the part of the user. Kinetikit is a user-friendly, click-and-drag interface for modeling models of chemical reactions such as occur in biochemical signaling pathways. In addition to defining and running kinetic models, it is intended to facilitate managing kinetic data in these complex models (Bhalla, 1998; Bhalla and Iyengar, 1999).
GENESIS comes with extensive documentation. The GENESIS Reference Manual comes in three forms: a 566 page manual in postscript format, and corresponding on-line help available as either plain text files viewable within the simulator, or hypertext help, which may viewed with a web browser.
To complement the Reference Manual, we have published two editions of The Book of GENESIS. The most recent edition (Bower and Beeman, 1998) contains a CD-ROM with the GENESIS distribution, documentation, and files from the Users Group archives. It is being widely used in both research and teaching, and consists of two parts serving complementary needs. Part I is designed to supplement instruction in neurobiology within upper division undergraduate and graduate neuroscience courses and contains chapters on various topics, each written by a known expert to accompany a particular GENESIS tutorial. These interactive tutorial simulations are user-friendly with on-line help and may be used without any prior knowledge of the GENESIS simulator or computer programming. Part II serves as a users guide to GENESIS, complementing the GENESIS Reference Manual, by introducing the basic features of GENESIS as well as the process of creating GENESIS simulations, providing a starting point for the development of new simulations. (For further details, please see http://www.genesis-sim.org/GENESIS/bog/bog.html.)
The tutorials mentioned above are included in the GENESIS distribution, along with other tutorials and demonstrations designed to aid new users in building GENESIS simulations. These illustrate the use of advanced GENESIS features, including: objects for modeling calcium diffusion; objects for spike analysis, recording and generation; the use of facilitating and Hebbian synapses; objects for modeling stochastic ion channels; and the parameter search library. Several of the tutorials are based on significant published research simulations including the piriform cortex (Wilson and Bower, 1992), the hippocampal pyramidal cell (Traub, et al., 1991), and the detailed cerebellar Purkinje cell model (De Schutter and Bower, 1994a,b). In addition to their educational purpose, these tutorial simulations provide examples of well-constructed GENESIS simulations, and have been used by others as the basis for the construction of new published research simulations.
Individuals or research groups who make serious use of GENESIS are encouraged to join the GENESIS Users Group, BABEL. There are currently 347 BABEL memberships, representing approximately 700 users. Members of BABEL are entitled to access the BABEL directories and participate in the email newsgroup. These are used as a repository for the latest contributions by GENESIS users and developers. These include new simulations, libraries of cells and channels, additional simulator components, new documentation and tutorials, bug reports and fixes, and the posting of questions and hints for setting up GENESIS simulations. As the results of GENESIS research simulations are published, many of these simulations are being made available through BABEL.
From its inception, GENESIS has had a strong educational component. We are currently aware of 49 institutions in 11 countries that have used GENESIS in teaching. Many of these institutions use GENESIS in association with The Book of GENESIS (Bower and Beeman, 1998).
GENESIS and the tutorials described earlier are now being widely used in graduate and undergraduate instruction. These uses include full semester courses in computational neuroscience or neural modeling, short intensive courses or workshops, an option for a course project, and short units on computational neuroscience within courses on artificial neural nets. An example of the use of the GENESIS tutorials as the basis for a short unit on neural modeling is available on the GENESIS web site as an HTML version of two lectures given at the University of Colorado.
GENESIS has also formed the basis for the laboratory section of the Methods in Computational Neuroscience course (1988-1996) at the Marine Biological Laboratory, a course in Mexico City in the summer of 1991, the Crete course in Computational Neuroscience (1996-1998), the EU Advanced Course in Computational Neuroscience (1999-2002), and the Computational Neuroscience course at the National Centre for Biological Sciences in Bangalore (1999-2000).
In the many years since the GENESIS project began at Caltech, its development has spread to many other institutions including the University of Texas at San Antonio, the University of Antwerp, the National Centre for Biological Studies in Bangalore, the University of Colorado, the Pittsburgh Supercomputer Center, the San Diego Supercomputer Center, and Emory University. In addition, the tools available for graphical user interfaces, and the decentralization of computational resources due to the widespread use of the World Wide Web, have dramatically changed the environment for computer-based education and research. In a collaborative effort involving many institutions, we are currently redesigning and reimplementing GENESIS in order to modernize the user interface and link the process of modeling with online databases of models and model components. Our efforts are taking place in the context of a new software framework called the Modeler's Workspace (Forss et al,. 1999; Hucka et al., 2002) and a simulator-independent representation of neural models called NeuroML (Goddard, et al., 2001).
Further information about GENESIS and PGENESIS, as well as instructions for download and installation may be obtained from the GENESIS web site, http://www.genesis-sim.org/GENESIS/. Inquiries concerning GENESIS should be addressed to genesis@genesis-sim.org.
Bhalla, U.S. and Iyengar, R., 1999, Emergent properties of networks of biological signaling pathways, Science 283:381-387.
* Bower, J.M., 1995, Reverse engineering the nervous system: An in vivo, in vitro, and in computo approach to understanding the mammalian olfactory system, in An Introduction to Neural and Electronic Networks, (S. F. Zornetzer, J. L. Davis and C. Lau Eds.), second edn, New York: Academic Press, pp. 3-28.
* Bower, J.M., and Beeman, D., 1998, The Book of GENESIS: Exploring Realistic Neural Models with the GEneral NEural SImulation System, second edition, New York: Springer-Verlag.
De Schutter, E., and Beeman, D., 1998, Speeding Up GENESIS Simulations, in The Book of GENESIS: Exploring Realistic Neural Models with the GEneral NEural SImulation System, (J.M. Bower and D. Beeman, Eds.), second edition, New York: Springer-Verlag. pp. 329-447.
De Schutter, E., and Bower, J.M., 1994a, An active membrane model of the cerebellar Purkinje cell. I. Simulation of current clamps in slice, J. Neurophysiol., 71:375-400.
De Schutter, E., and Bower, J.M., 1994b. An active membrane model of the cerebellar Purkinje cell. II. Simulation of synaptic responses, J. Neurophysiol., 71:401-419.
* Forss, J., Beeman, D., Eickler-West, R., and Bower, J.M., 1999, The Modeler's Workspace: A Distributed Digital Library for Neuroscience, Future Generation Computer Systems, 16:111-121.
* Goddard, N., Hucka, M., Howell, F., Cornelis, H., Shankar, K., and Beeman, D., 2001, ``Towards NeuroML: Model Description Methods for Collaborative Modelling in Neuroscience'', Phil. Trans. R. Soc. Lond. B, 356:1209-1228.
* Hucka , M., Shankar, K., Beeman, D., and Bower, J.M., 2002, The Modeler's Workspace: Making model-based studies of the nervous system more accessible, in Computational Neuroanatomy: Principles and Methods, (G. Ascoli, Ed.), Totowa, NJ: Humana Press.
Stricanne, B., Morissette, J., and Bower, J.M., 1998, Exploring the sources of cerebellar post-lesion plasticity with a network model of the somatosensory system, Soc. Neurosci. Abs. 23:2364.
Traub, R.D., Wong, R.K.S., Miles, R., and Michelson, H., 1991, A model of a CA3 hippocampal pyramidal neuron incorporating voltage-clamp data on intrinsic conductances, J. Neurophysiol., 66:635-650.
Wilson, M., and Bower, J.M., 1992, Simulating cerebral cortical networks: oscillations and temporal interactions in a computer simulation of piriform (olfactory) cortex, J. Neurophysiol., 67:981-995.